02-cairns- atul
Asian J. Exp. Sci., Vol. 22, No. 1, 2008; 143-146 Concurrent Effects of Eyestalk Ablation and Fluoxetine on the Nutrient Depostion During Ovarian Development in a Fresh Water Prawn, Machrobrachium lamarrei lamarrei R. Eswaralakshmi, J. Jayanthi and M.G. Ragunathan Department of Advanced Zoology and Biotechnology, Guru Nanak College, Abstract : The organic compounds like protein, carboh

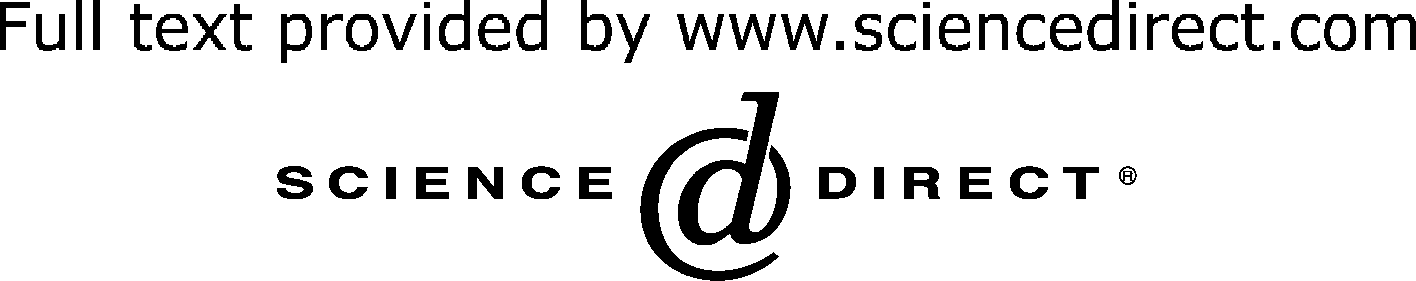 Chemical biology and bacteria: not simply a matter oflife or deathDeborah T and Eric J Rubin
Chemical biological approaches to understanding bacteria
small molecules as tools to systematically dissect the
have largely been confined to screening for antibiotics. More
pathways involved in these complex phenotypes, culmi-
complex phenotypes, such as virulence, have largely been
nating in the generation of a new name for an old
studied using bacterial genetics. However, it has recently
become clear that these two methods are complementary andthat combining chemical biologic and genetic approaches to
The field of chemical genetics has predominantly fallen
studying bacteria brings new power to old problems.
Chemical biology and bacteria: not simply a matter oflife or deathDeborah T and Eric J Rubin
Chemical biological approaches to understanding bacteria
small molecules as tools to systematically dissect the
have largely been confined to screening for antibiotics. More
pathways involved in these complex phenotypes, culmi-
complex phenotypes, such as virulence, have largely been
nating in the generation of a new name for an old
studied using bacterial genetics. However, it has recently
become clear that these two methods are complementary andthat combining chemical biologic and genetic approaches to
The field of chemical genetics has predominantly fallen
studying bacteria brings new power to old problems.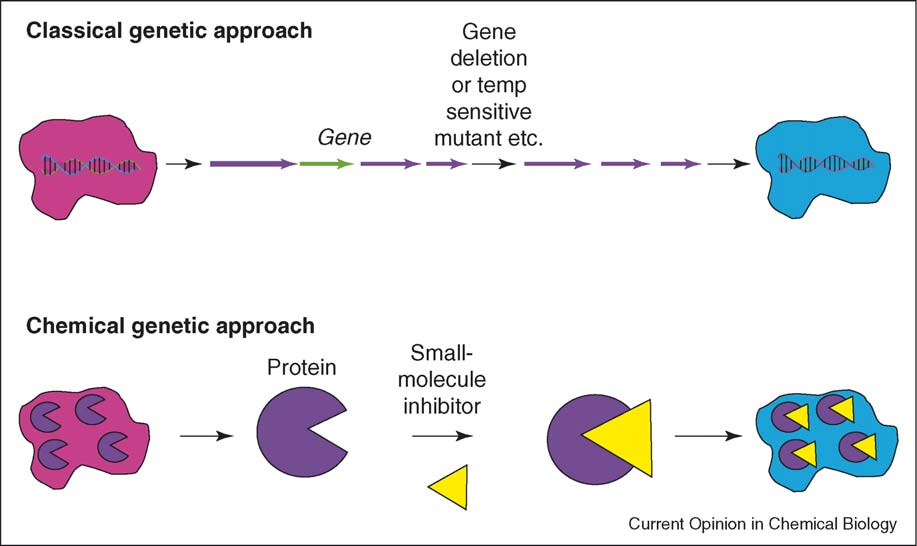 Comparison of classical and chemical genetics
Slow (except for conditionally stable proteins)
Useful for studying proteins essential for in vitro survival
By analogy to classical genetics, both forward and reverse
expensive and more robust, having been performed in
genetic strategies may be employed in high-throughput
diverse organisms including Yersinia, Vibrio cholerae, Sta-
chemical screens to find small molecules of interest
phylococcus aureus and Escherichia coli [].
Comparison of classical and chemical genetics
Slow (except for conditionally stable proteins)
Useful for studying proteins essential for in vitro survival
By analogy to classical genetics, both forward and reverse
expensive and more robust, having been performed in
genetic strategies may be employed in high-throughput
diverse organisms including Yersinia, Vibrio cholerae, Sta-
chemical screens to find small molecules of interest
phylococcus aureus and Escherichia coli [].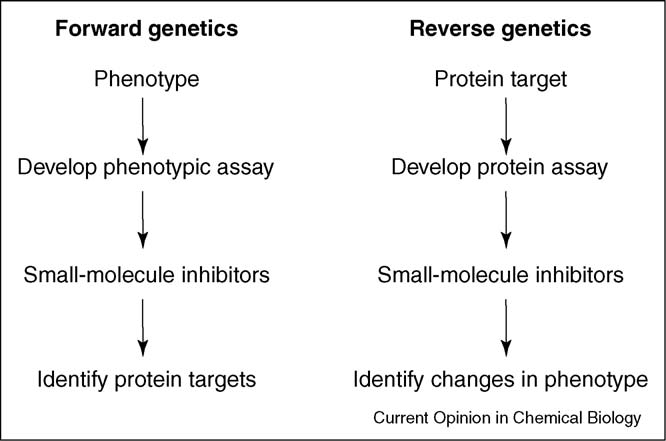 Chemical biology and bacteria: not simply a matter of life or death Hung and Rubin
proteins whose function is lost at high temperature. Suchmutants may be problematic as they grow only underrestricted conditions that can result in numerous unre-lated changes to the cell, and the mutant proteins oftenlack full activity under permissive conditions. Moreimportantly, temperature-sensitive mutants can only beisolated by a relatively laborious process that is onlysuccessful for about a third of known essential genes [
The limitation of these types of traditional mutants is thatthey are not truly conditional. One cannot turn the geneproducts on and off at will on a short time scale, in atunable manner that is also reversible. This limitation hasseveral implications. Essential genes are difficult to studybecause it is difficult to isolate mutations that result inlethality. Generation of mutations on the DNA leveloften results in compensatory changes such as up-regula-
Forward and reverse approaches to using chemical genetics.
Chemical biology and bacteria: not simply a matter of life or death Hung and Rubin
proteins whose function is lost at high temperature. Suchmutants may be problematic as they grow only underrestricted conditions that can result in numerous unre-lated changes to the cell, and the mutant proteins oftenlack full activity under permissive conditions. Moreimportantly, temperature-sensitive mutants can only beisolated by a relatively laborious process that is onlysuccessful for about a third of known essential genes [
The limitation of these types of traditional mutants is thatthey are not truly conditional. One cannot turn the geneproducts on and off at will on a short time scale, in atunable manner that is also reversible. This limitation hasseveral implications. Essential genes are difficult to studybecause it is difficult to isolate mutations that result inlethality. Generation of mutations on the DNA leveloften results in compensatory changes such as up-regula-
Forward and reverse approaches to using chemical genetics.