Massage and herbal medicine news
p: 02 94360306 e: [email protected] w: www.pennysnaturalhealth.biz Massage and herbal medicine news Father's Day This year Father’ Day is Sunday 5 September. This is a day for Dads’ to be celebrated for their contribution to family. I have gift cards which can be used for massage to help the males in your life to relax or to help them with a specific pro
 Small Metallothionein MT-10 Genes in Coastal and HydrothermalMussels
Laboratoire de Biologic et Ge´ne´tique Evolutive, Universite´ du Maine, 72085 Le Mans, France
Received: 25 November 2003 / Accepted: 20 August 2004 / Online publication: 5 May 2005
motif: Cys-Cys, Cys-X-Cys, and Cys-X-X-Cys (whereX is an amino acid other than cysteine). Mollusk
Metallothioneins (MTs) are important proteins in
MTs all belong to class I, as do mammalian and
the intracellular regulation of metals. In the Mytil-
crustacean MTs (Kagi, 1993; Picinni et al., 1999).
Small Metallothionein MT-10 Genes in Coastal and HydrothermalMussels
Laboratoire de Biologic et Ge´ne´tique Evolutive, Universite´ du Maine, 72085 Le Mans, France
Received: 25 November 2003 / Accepted: 20 August 2004 / Online publication: 5 May 2005
motif: Cys-Cys, Cys-X-Cys, and Cys-X-X-Cys (whereX is an amino acid other than cysteine). Mollusk
Metallothioneins (MTs) are important proteins in
MTs all belong to class I, as do mammalian and
the intracellular regulation of metals. In the Mytil-
crustacean MTs (Kagi, 1993; Picinni et al., 1999). V. LEIGNEL ET AL.: SMALL METALLOTHIONEIN GENES IN MUSSELS
Fig. 1. Multiple alignments of MT-10 cDNA metallothionein available in GenBank with small MT-10 genes isolated fromMytilus edulis (MT-10A and MT-10B) and Bathymodiolus thermophilus (MT-10Bt). The GenBank accession numbers ofthe cDNA sequences are as follows: MT-10III (AJ005454), MT-10II (AJ005453), MT-10Ib (AJ005452), MT-10Ia (AJ005451),MT-10IV (AJ007506). Dots indicate deletions. The motif corresponding to MT-10I and MT-10II is shown in bold faceletters. The specific insertion and deletion sites for MT-10A and MT-10B (the MT-10 genes of Mytilus edulis) are enclosedwith dashed and solid boxes, respectively. The specific sites of the MT-10Bt sequences in Bathymodiolus thermophilusare indicated by italics and underlining.
V. LEIGNEL ET AL.: SMALL METALLOTHIONEIN GENES IN MUSSELS
Fig. 1. Multiple alignments of MT-10 cDNA metallothionein available in GenBank with small MT-10 genes isolated fromMytilus edulis (MT-10A and MT-10B) and Bathymodiolus thermophilus (MT-10Bt). The GenBank accession numbers ofthe cDNA sequences are as follows: MT-10III (AJ005454), MT-10II (AJ005453), MT-10Ib (AJ005452), MT-10Ia (AJ005451),MT-10IV (AJ007506). Dots indicate deletions. The motif corresponding to MT-10I and MT-10II is shown in bold faceletters. The specific insertion and deletion sites for MT-10A and MT-10B (the MT-10 genes of Mytilus edulis) are enclosedwith dashed and solid boxes, respectively. The specific sites of the MT-10Bt sequences in Bathymodiolus thermophilusare indicated by italics and underlining.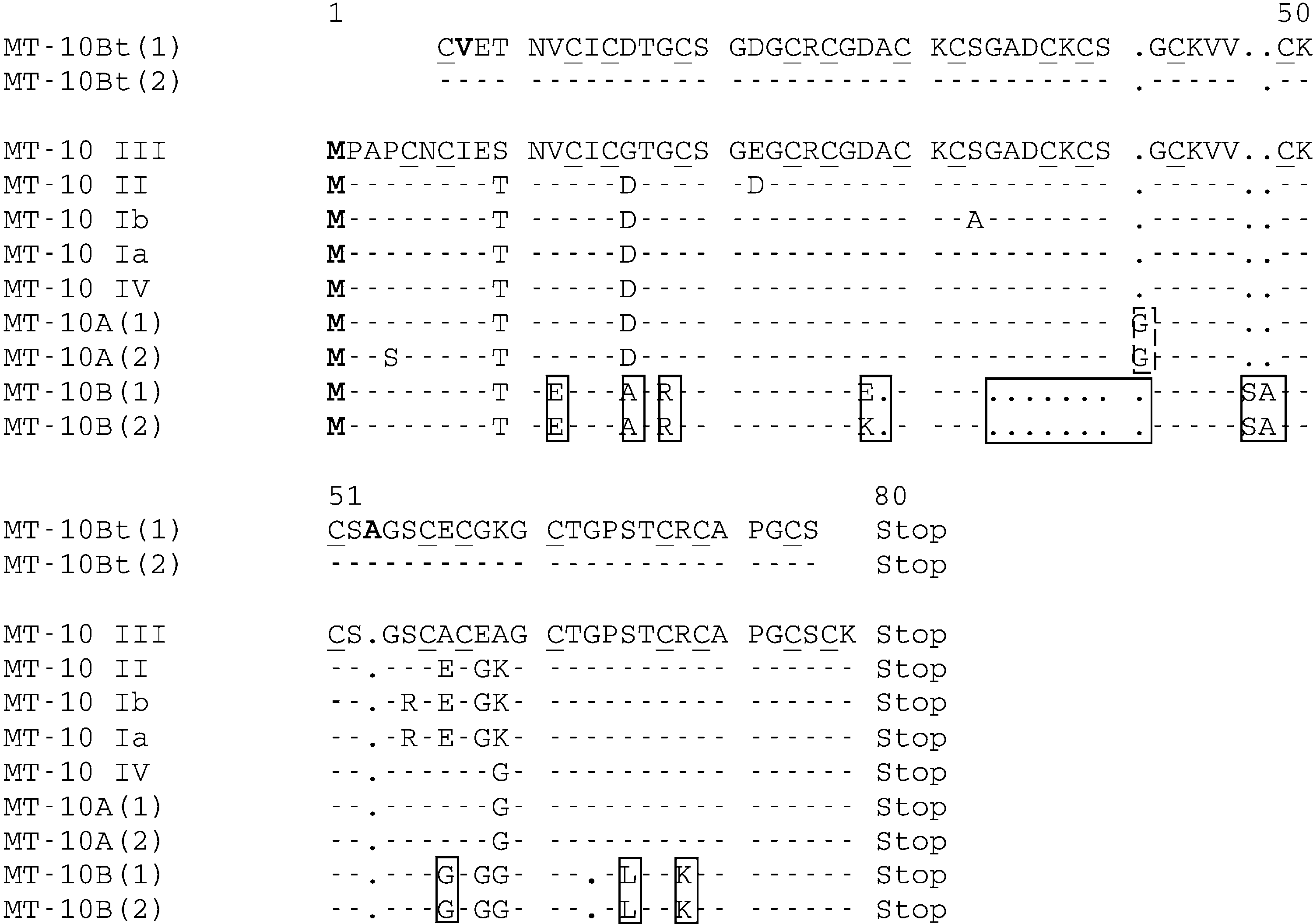 V. LEIGNEL ET AL.: SMALL METALLOTHIONEIN GENES IN MUSSELS
Fig. 2. Multiple alignments of a protein sequence obtained in silico (bioinformatic approach) from the MT-10 nucleicsequence (showed in the Figure 1) obtained from Mytitus edulis and Bathymodiolus thermophilus. The GenBankaccession numbers of the cDNA sequences are as follows: MT-10III (AJ005454), MT-10II (AJ005453), MT-10Ib (AJ005452),MT-10Ia (AJ005451), and MT-10IV (AJ007506). The cysteine residues characteristic of metallothioneins are underlined.
V. LEIGNEL ET AL.: SMALL METALLOTHIONEIN GENES IN MUSSELS
Fig. 2. Multiple alignments of a protein sequence obtained in silico (bioinformatic approach) from the MT-10 nucleicsequence (showed in the Figure 1) obtained from Mytitus edulis and Bathymodiolus thermophilus. The GenBankaccession numbers of the cDNA sequences are as follows: MT-10III (AJ005454), MT-10II (AJ005453), MT-10Ib (AJ005452),MT-10Ia (AJ005451), and MT-10IV (AJ007506). The cysteine residues characteristic of metallothioneins are underlined.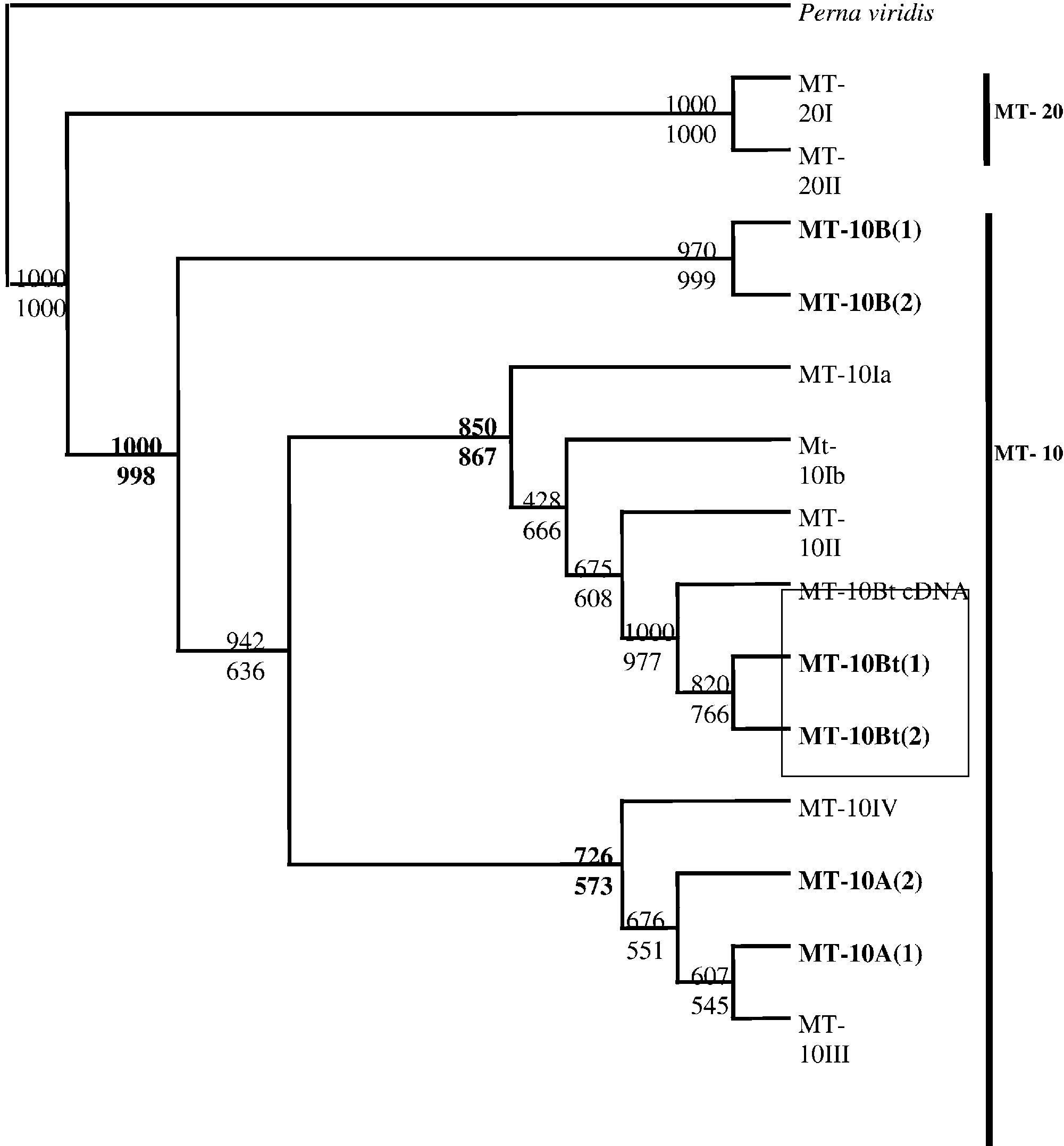 V. LEIGNEL ET AL.: SMALL METALLOTHIONEIN GENES IN MUSSELS
Wegnez, 1995). In Crassostrea gigas, another atypi-cal MT gene, CgMT2, which has an organizationcharacterized by an exon duplication, has been iso-lated and this also seems to be an active gene (Tan-guy and Moraga, 2001; Tanguy et al., 2001).
V. LEIGNEL ET AL.: SMALL METALLOTHIONEIN GENES IN MUSSELS
Wegnez, 1995). In Crassostrea gigas, another atypi-cal MT gene, CgMT2, which has an organizationcharacterized by an exon duplication, has been iso-lated and this also seems to be an active gene (Tan-guy and Moraga, 2001; Tanguy et al., 2001).